Data engineering
With Data preparation tutorial, we prepared datasets (geospatial time-series data) to analyze. As the next step of data engineering, we will perform the followings here.
Data cleaning
Data transformation
Arithmetic operations
EDA at a geospatial layer
Data subsetting for a location and data complement
EDA of subset
Note that EDA = explanatory data analysis
[1]:
from pprint import pprint
import covsirphy as cs
import numpy as np
cs.__version__
[1]:
'3.1.2.delta'
We will use the recommended datasets at country-level data as an example.
[2]:
eng = cs.DataEngineer()
eng.download(databases=["japan", "covid19dh", "owid"])
eng.all().info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 298052 entries, 0 to 298051
Data columns (total 27 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ISO3 298052 non-null category
1 Province 298052 non-null category
2 City 298052 non-null category
3 Date 298052 non-null datetime64[ns]
4 Cancel_events 197839 non-null Float64
5 Confirmed 239411 non-null Float64
6 Contact_tracing 197865 non-null Float64
7 Country 287385 non-null string
8 Fatal 222214 non-null Float64
9 Gatherings_restrictions 197839 non-null Float64
10 Information_campaigns 197865 non-null Float64
11 Internal_movement_restrictions 197865 non-null Float64
12 International_movement_restrictions 197872 non-null Float64
13 Population 286223 non-null Float64
14 Product 176264 non-null string
15 Recovered 74399 non-null Float64
16 School_closing 197864 non-null Float64
17 Stay_home_restrictions 197833 non-null Float64
18 Stringency_index 197828 non-null Float64
19 Testing_policy 197865 non-null Float64
20 Tests 91191 non-null Float64
21 Transport_closing 197845 non-null Float64
22 Vaccinated_full 59857 non-null Float64
23 Vaccinated_once 62933 non-null Float64
24 Vaccinations 66925 non-null Float64
25 Vaccinations_boosters 38050 non-null Float64
26 Workplace_closing 197864 non-null Float64
dtypes: Float64(21), category(3), datetime64[ns](1), string(2)
memory usage: 61.7 MB
1. Data cleaning
DataEngineer().clean() performs the following data cleaning functionalities. By applying a list of strings to kinds argument (eg. kinds=["resample"]), we can specify the cleaning method(s).
“convert_date”: Convert dtype of date column to pandas.Timestamp.
“resample”: Resample records with dates.
“fillna”: Fill NA values with ‘-’ (layers) and the previous values and 0.
For “convert_date”, keyword arguments of pandas.to_datetime() including “dayfirst (bool): whether date format is DD/MM or not” can be used.
For “resample”, date_range=<tuple of (str or None, str or None) or None>) can be applied as keyword arguments to set the range.
[3]:
eng.clean()
eng.all().info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 298667 entries, 0 to 298666
Data columns (total 27 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ISO3 298667 non-null category
1 Province 298667 non-null category
2 City 298667 non-null category
3 Date 298667 non-null datetime64[ns]
4 Contact_tracing 298667 non-null Float64
5 Vaccinated_once 298667 non-null Float64
6 Confirmed 298667 non-null Float64
7 Country 298667 non-null object
8 Stay_home_restrictions 298667 non-null Float64
9 Vaccinated_full 298667 non-null Float64
10 Vaccinations 298667 non-null Float64
11 International_movement_restrictions 298667 non-null Float64
12 Gatherings_restrictions 298667 non-null Float64
13 Stringency_index 298667 non-null Float64
14 Transport_closing 298667 non-null Float64
15 Testing_policy 298667 non-null Float64
16 Recovered 298667 non-null Float64
17 Fatal 298667 non-null Float64
18 Internal_movement_restrictions 298667 non-null Float64
19 Workplace_closing 298667 non-null Float64
20 Cancel_events 298667 non-null Float64
21 Population 298667 non-null Float64
22 Tests 298667 non-null Float64
23 Vaccinations_boosters 298667 non-null Float64
24 School_closing 298667 non-null Float64
25 Information_campaigns 298667 non-null Float64
26 Product 298667 non-null object
dtypes: Float64(21), category(3), datetime64[ns](1), object(2)
memory usage: 61.8+ MB
2. Data transformation
Transform all registered data, calculating the number of susceptible and infected cases. This is required to analyze real data with SIR-derived models.
Susceptible = Population - Confirmed
Infected = Confirmed - Fatal - Recovered
[4]:
main_variables = ["Population", "Susceptible", "Confirmed", "Infected", "Fatal", "Recovered"]
[5]:
eng.transform()
eng.all(variables=main_variables).tail()
[5]:
| ISO3 | Province | City | Date | Population | Susceptible | Confirmed | Infected | Fatal | Recovered | |
|---|---|---|---|---|---|---|---|---|---|---|
| 298662 | ZWE | - | - | 2023-03-05 | 14439018.0 | 14174891.0 | 264127.0 | 175465.0 | 5668.0 | 82994.0 |
| 298663 | ZWE | - | - | 2023-03-06 | 14439018.0 | 14174891.0 | 264127.0 | 175465.0 | 5668.0 | 82994.0 |
| 298664 | ZWE | - | - | 2023-03-07 | 14439018.0 | 14174891.0 | 264127.0 | 175465.0 | 5668.0 | 82994.0 |
| 298665 | ZWE | - | - | 2023-03-08 | 14439018.0 | 14174742.0 | 264276.0 | 175611.0 | 5671.0 | 82994.0 |
| 298666 | ZWE | - | - | 2023-03-09 | 14439018.0 | 14174742.0 | 264276.0 | 175611.0 | 5671.0 | 82994.0 |
Recalculation of “Population” and “Confirmed” can be performed with DataEngineer().inverse_transform(), if necessary. (No impact with this example data.)
[6]:
eng.inverse_transform()
eng.all(variables=main_variables).tail()
[6]:
| ISO3 | Province | City | Date | Population | Susceptible | Confirmed | Infected | Fatal | Recovered | |
|---|---|---|---|---|---|---|---|---|---|---|
| 298662 | ZWE | - | - | 2023-03-05 | 14439018.0 | 14174891.0 | 264127.0 | 175465.0 | 5668.0 | 82994.0 |
| 298663 | ZWE | - | - | 2023-03-06 | 14439018.0 | 14174891.0 | 264127.0 | 175465.0 | 5668.0 | 82994.0 |
| 298664 | ZWE | - | - | 2023-03-07 | 14439018.0 | 14174891.0 | 264127.0 | 175465.0 | 5668.0 | 82994.0 |
| 298665 | ZWE | - | - | 2023-03-08 | 14439018.0 | 14174742.0 | 264276.0 | 175611.0 | 5671.0 | 82994.0 |
| 298666 | ZWE | - | - | 2023-03-09 | 14439018.0 | 14174742.0 | 264276.0 | 175611.0 | 5671.0 | 82994.0 |
3. Arithmetic operations
We can perform arithmetic operations to add new columns.
.diff(column, suffix="_diff", freq="D"): Calculate daily new cases with “f(x>0) = F(x) - F(x-1), x(0) = 0 when F is cumulative numbers”..add(columns, new=None, fill_value=0): Calculate element-wise addition with pandas.DataFrame.sum(axis=1), X1 + X2 + X3 +….mul(columns, new=None, fill_value=0): Calculate element-wise multiplication with pandas.DataFrame.product(axis=1), X1 * X2 * X3 *….sub(minuend, subtrahend, new=None, fill_value=0): Calculate element-wise subtraction with pandas.Series.sub(), minuend - subtrahend..div(columns, new=None, fill_value=0): Calculate element-wise floating division with pandas.Series.div(), numerator / denominator..assign(**kwargs)): Assign a new column with pandas.DataFrame.assign().
[7]:
# Diff
eng.diff(column="Confirmed", suffix="_diff", freq="D")
eng.all(variables=["Confirmed", "Confirmed_diff"]).tail()
[7]:
| ISO3 | Province | City | Date | Confirmed | Confirmed_diff | |
|---|---|---|---|---|---|---|
| 298662 | ZWE | - | - | 2023-03-05 | 264127.0 | 0.0 |
| 298663 | ZWE | - | - | 2023-03-06 | 264127.0 | 0.0 |
| 298664 | ZWE | - | - | 2023-03-07 | 264127.0 | 0.0 |
| 298665 | ZWE | - | - | 2023-03-08 | 264276.0 | 149.0 |
| 298666 | ZWE | - | - | 2023-03-09 | 264276.0 | 0.0 |
[8]:
# Addition
eng.add(columns=["Fatal", "Recovered"])
eng.all(variables=["Fatal", "Recovered", "Fatal+Recovered"]).tail()
[8]:
| ISO3 | Province | City | Date | Fatal | Recovered | Fatal+Recovered | |
|---|---|---|---|---|---|---|---|
| 298662 | ZWE | - | - | 2023-03-05 | 5668.0 | 82994.0 | 88662.0 |
| 298663 | ZWE | - | - | 2023-03-06 | 5668.0 | 82994.0 | 88662.0 |
| 298664 | ZWE | - | - | 2023-03-07 | 5668.0 | 82994.0 | 88662.0 |
| 298665 | ZWE | - | - | 2023-03-08 | 5671.0 | 82994.0 | 88665.0 |
| 298666 | ZWE | - | - | 2023-03-09 | 5671.0 | 82994.0 | 88665.0 |
[9]:
# Multiplication
eng.mul(columns=["Confirmed", "Recovered"])
eng.all(variables=["Confirmed", "Recovered", "Confirmed*Recovered"]).tail()
[9]:
| ISO3 | Province | City | Date | Confirmed | Recovered | Confirmed*Recovered | |
|---|---|---|---|---|---|---|---|
| 298662 | ZWE | - | - | 2023-03-05 | 264127.0 | 82994.0 | 21920956238.0 |
| 298663 | ZWE | - | - | 2023-03-06 | 264127.0 | 82994.0 | 21920956238.0 |
| 298664 | ZWE | - | - | 2023-03-07 | 264127.0 | 82994.0 | 21920956238.0 |
| 298665 | ZWE | - | - | 2023-03-08 | 264276.0 | 82994.0 | 21933322344.0 |
| 298666 | ZWE | - | - | 2023-03-09 | 264276.0 | 82994.0 | 21933322344.0 |
[10]:
# Division
eng.div(numerator="Confirmed", denominator="Tests", new="Positive_rate")
# Assignment of new a new column
eng.assign(**{"Positive_rate_%": lambda x: x["Positive_rate"] * 100})
eng.all(variables=["Tests", "Confirmed", "Positive_rate_%"]).tail()
[10]:
| ISO3 | Province | City | Date | Tests | Confirmed | Positive_rate_% | |
|---|---|---|---|---|---|---|---|
| 298662 | ZWE | - | - | 2023-03-05 | 2379907.0 | 264127.0 | 11.098207 |
| 298663 | ZWE | - | - | 2023-03-06 | 2379907.0 | 264127.0 | 11.098207 |
| 298664 | ZWE | - | - | 2023-03-07 | 2379907.0 | 264127.0 | 11.098207 |
| 298665 | ZWE | - | - | 2023-03-08 | 2379907.0 | 264276.0 | 11.104468 |
| 298666 | ZWE | - | - | 2023-03-09 | 2379907.0 | 264276.0 | 11.104468 |
4. EDA at a geospatial layer
DataEngineer().layer() returns the data at the selected layer in the date range.
Arguments:
geo (tuple(list[str] or tuple(str) or str) or str or None): location names to specify the layer or None (the top level)
start_date (str or None): start date, like 22Jan2020
end_date (str or None): end date, like 01Feb2020
variables (list[str] or None): list of variables to add or None (all available columns)
[11]:
eng.layer().tail()
[11]:
| ISO3 | Province | City | Date | Cancel_events | Confirmed | Confirmed*Recovered | Confirmed_diff | Contact_tracing | Country | ... | Stringency_index | Susceptible | Testing_policy | Tests | Transport_closing | Vaccinated_full | Vaccinated_once | Vaccinations | Vaccinations_boosters | Workplace_closing | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 298662 | ZWE | - | - | 2023-03-05 | 1.0 | 264127.0 | 21920956238.0 | 0.0 | 1.0 | Zimbabwe | ... | 53.7 | 14174891.0 | 3.0 | 2379907.0 | 1.0 | 4751270.0 | 6437808.0 | 12222754.0 | 1033676.0 | 1.0 |
| 298663 | ZWE | - | - | 2023-03-06 | 1.0 | 264127.0 | 21920956238.0 | 0.0 | 1.0 | Zimbabwe | ... | 53.7 | 14174891.0 | 3.0 | 2379907.0 | 1.0 | 4751270.0 | 6437808.0 | 12222754.0 | 1033676.0 | 1.0 |
| 298664 | ZWE | - | - | 2023-03-07 | 1.0 | 264127.0 | 21920956238.0 | 0.0 | 1.0 | Zimbabwe | ... | 53.7 | 14174891.0 | 3.0 | 2379907.0 | 1.0 | 4751270.0 | 6437808.0 | 12222754.0 | 1033676.0 | 1.0 |
| 298665 | ZWE | - | - | 2023-03-08 | 1.0 | 264276.0 | 21933322344.0 | 149.0 | 1.0 | Zimbabwe | ... | 53.7 | 14174742.0 | 3.0 | 2379907.0 | 1.0 | 4751270.0 | 6437808.0 | 12222754.0 | 1033676.0 | 1.0 |
| 298666 | ZWE | - | - | 2023-03-09 | 1.0 | 264276.0 | 21933322344.0 | 0.0 | 1.0 | Zimbabwe | ... | 53.7 | 14174742.0 | 3.0 | 2379907.0 | 1.0 | 4751270.0 | 6437808.0 | 12222754.0 | 1033676.0 | 1.0 |
5 rows × 34 columns
This dataset has only country-level data and geo should be country name. We can select the followings as geo argument for EDA at a geospatial layer when we have adequate data.
When
geo=Noneorgeo=(None,), returns country-level data, assuming we have country/province/city as layers here.When
geo=("Japan",)orgeo="Japan", returns province-level data in Japan.When
geo=(["Japan", "UK"],), returns province-level data in Japan and UK.When
geo=("Japan", "Kanagawa"), returns city-level data in Kanagawa/Japan.When
geo=("Japan", ["Tokyo", "Kanagawa"]), returns city-level data in Tokyo/Japan and Kanagawa/Japan.
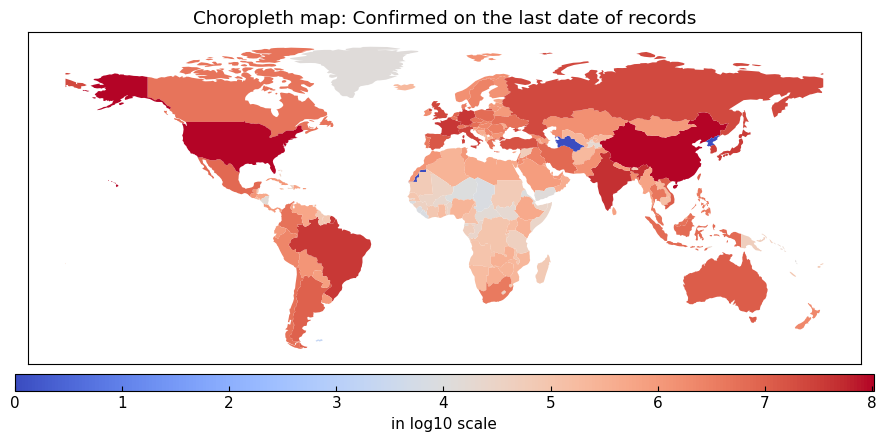
Additionally, we can create a choropleth map with a data at a geospatial layer on a date.
arguments of DataEngineer().choropleth():
geo (tuple(list[str] or tuple(str) or str) or str or None): location names to specify the layer or None (the top level)
variable (str): variable name to show
on (str or None): the date, like 22Jan2020, or None (the last date of each location)
title (str): title of the map
filename (str or None): filename to save the figure or None (display)
logscale (bool): whether convert the value to log10 scale values or not
directory (str): directory to save GeoJSON file of “Natural Earth” GitHub repository
natural_earth (str or None): title of GeoJSON file (without extension) of “Natural Earth” GitHub repository or None (automatically determined)
**kwargs: keyword arguments of the following classes and methods.
matplotlib.pyplot.savefig(), matplotlib.pyplot.legend(), and
pandas.DataFrame.plot()
[12]:
eng.choropleth(geo=None, variable="Confirmed", title="Choropleth map: Confirmed on the last date of records", filename=None)
5. Data subsetting for a location and data complement
The dataset is a geospatial time-series data. By selecting a location, the dataset will be converted to a time-series data, which is easier to analyze.
5.1 Subsetting
We will create a subset for selected location (eg. country, province/prefecture/state, city). Because the loaded dataset has country-level data, total values in United Kingdom (UK) on dates will be created here as an example.
Arguments of DataEngineer().subset():
geo (tuple(list[str] or tuple(str) or str) or str or None): location names to filter or None (total at the top level)
start_date (str or None): start date, like 22Jan2020
end_date (str or None): end date, like 01Feb2020
variables (list[str] or None): list of variables to add or None (all available columns)
complement (bool): whether perform data complement or not, True as default
get_dummies (bool): whether convert categorical variable into dummy variables or not, True as default
**Kwargs: keyword arguments for complement and default values
recovery_period (int): expected value of recovery period [days], 17
interval (int): expected update interval of the number of recovered cases [days], 2
max_ignored (int): Max number of recovered cases to be ignored [cases], 100
max_ending_unupdated (int): Max number of days to apply full complement, where max recovered cases are not updated [days], 14
upper_limit_days (int): maximum number of valid partial recovery periods [days], 90
lower_limit_days (int): minimum number of valid partial recovery periods [days], 7
upper_percentage (float): fraction of partial recovery periods with value greater than upper_limit_days, 0.5
lower_percentage (float): fraction of partial recovery periods with value less than lower_limit_days, 0.5
geo argument for subsetting when we have adequate data.When
geo=Noneorgeo=(None,), returns global scale records (total values of all country-level data), assuming we have country/province/city as layers here.When
geo=("Japan",)orgeo="Japan", returns country-level data in Japan.When
geo=(["Japan", "UK"],), returns country-level data of Japan and UK.When
geo=("Japan", "Tokyo"), returns province-level data of Tokyo/Japan.When
geo=("Japan", ["Tokyo", "Kanagawa"]), returns total values of province-level data of Tokyo/Japan and Kanagawa/Japan.When
geo=("Japan", "Kanagawa", "Yokohama"), returns city-level data of Yokohama/Kanagawa/Japan.When
geo=(("Japan", "Kanagawa", ["Yokohama", "Kawasaki"]), returns total values of city-level data of Yokohama/Kanagawa/Japan and Kawasaki/Kanagawa/Japan.
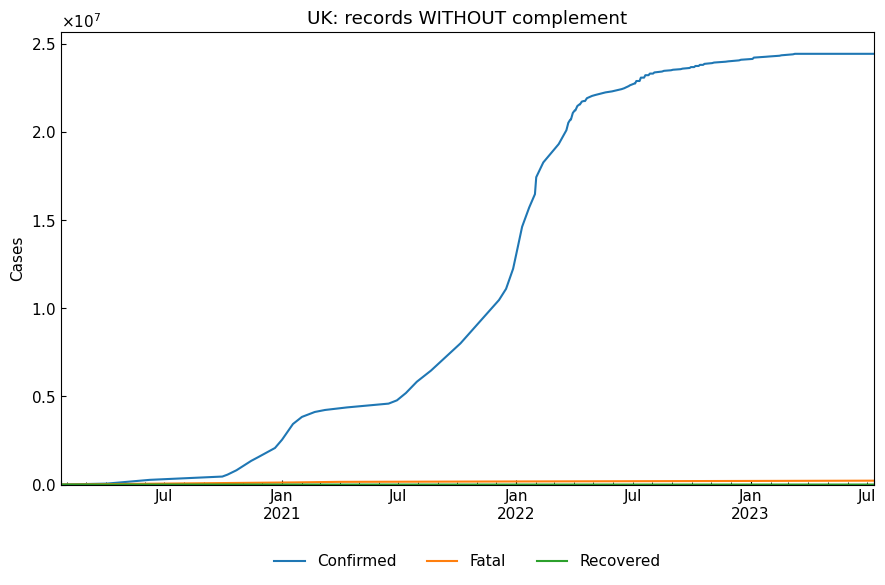
[13]:
# Without complement
without_df, status, status_dict = eng.subset(geo="UK", complement=False)
print(f"{status}\n")
pprint(status_dict)
cs.line_plot(without_df[["Confirmed", "Fatal", "Recovered"]], title="UK: records WITHOUT complement")
{}
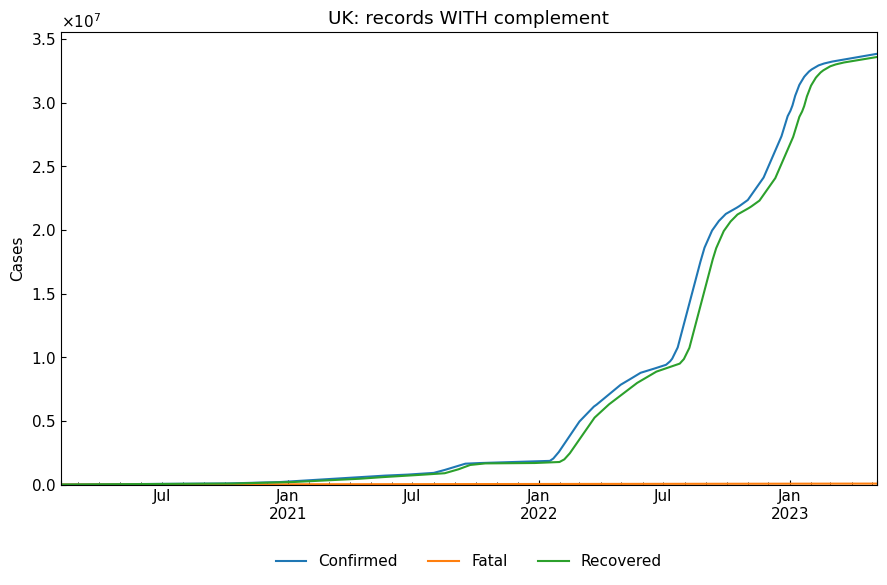
[14]:
# With complement (default)
with_df, status, status_dict = eng.subset(geo="Japan", complement=True)
print(f"{status}\n")
pprint(status_dict)
cs.line_plot(with_df[["Confirmed", "Fatal", "Recovered"]], title="UK: records WITH complement")
monotonic increasing complemented confirmed data and
monotonic increasing complemented fatal data and
fully complemented recovered data
{'Full_recovered': True,
'Monotonic_confirmed': True,
'Monotonic_fatal': True,
'Monotonic_recovered': True,
'Partial_recovered': False}
[15]:
with_df.info()
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 1189 entries, 2020-02-05 to 2023-05-08
Data columns (total 32 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Cancel_events 1189 non-null Float64
1 Confirmed*Recovered 1189 non-null Float64
2 Confirmed_diff 1189 non-null Float64
3 Contact_tracing 1189 non-null Float64
4 Fatal+Recovered 1189 non-null Float64
5 Gatherings_restrictions 1189 non-null Float64
6 Infected 1189 non-null Int64
7 Information_campaigns 1189 non-null Float64
8 Internal_movement_restrictions 1189 non-null Float64
9 International_movement_restrictions 1189 non-null Float64
10 Population 1189 non-null Float64
11 Positive_rate 1189 non-null Float64
12 Positive_rate_% 1189 non-null Float64
13 School_closing 1189 non-null Float64
14 Stay_home_restrictions 1189 non-null Float64
15 Stringency_index 1189 non-null Float64
16 Susceptible 1189 non-null Float64
17 Testing_policy 1189 non-null Float64
18 Tests 1189 non-null Float64
19 Transport_closing 1189 non-null Float64
20 Vaccinated_full 1189 non-null Float64
21 Vaccinated_once 1189 non-null Float64
22 Vaccinations 1189 non-null Float64
23 Vaccinations_boosters 1189 non-null Float64
24 Workplace_closing 1189 non-null Float64
25 Confirmed 1189 non-null Int64
26 Fatal 1189 non-null Int64
27 Recovered 1189 non-null Int64
28 Country_0 1189 non-null Int64
29 Country_Japan 1189 non-null Int64
30 Product_0 1189 non-null Int64
31 Product_Moderna, Novavax, Oxford/AstraZeneca, Pfizer/BioNTech 1189 non-null Int64
dtypes: Float64(24), Int64(8)
memory usage: 343.7 KB
5.2 Details of data complement
When complement=True (default), data complement will be performed for confirmed/fatal/recovered data. This may be required for analysis because reported cumulative values sometimes show decreasing by changing accidentally for an example. Additionally, some countries, including UK, do not report the number of recovered cases.
The possible kinds of complement for each country are the following:
“Monotonic_confirmed/fatal/recovered” (monotonic increasing complement) Force the variable show monotonic increasing.
“Full_recovered” (full complement of recovered data) Estimate the number of recovered cases using the value of estimated average recovery period.
“Partial_recovered” (partial complement of recovered data) When recovered values are not updated for some days, extrapolate the values.
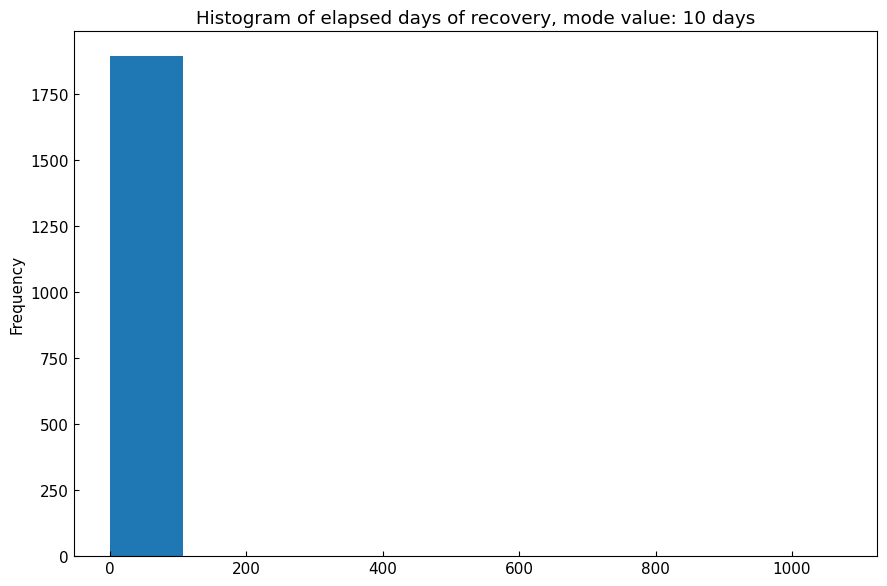
5.3 Recovery period
DataEngineer.recovery_period(data).[16]:
jpn_df, *_ = eng.subset(geo="Japan", variables=["Confirmed", "Fatal", "Recovered"], complement=False)
recovery_period = cs.DataEngineer.recovery_period(data=jpn_df)
print(f"Mode value of recovery period in Japan: {recovery_period} [days]")
Mode value of recovery period in Japan: 10 [days]
Details of recovery period calculation:
[17]:
df = jpn_df.resample("D").sum()
df["diff"] = df["Confirmed"] - df["Fatal"]
df = df.loc[:, ["diff", "Recovered"]].unstack().reset_index()
df.columns = ["Variable", "Date", "Number"]
df["Days"] = (df["Date"] - df["Date"].min()).dt.days
df = df.pivot_table(values="Days", index="Number", columns="Variable")
df = df.interpolate(limit_area="inside").dropna().astype(np.int64)
df["Elapsed"] = df["Recovered"] - df["diff"]
df = df.loc[df["Elapsed"] > 0]
# Calculate mode value
mode_value = round(df["Elapsed"].mode().mean())
df["Elapsed"].plot.hist(title=f"Histogram of elapsed days of recovery, mode value: {mode_value} days");
5.4 Alias of subsets
We can register alias names of subsets with DataEngineer().subset_alias().
Arguments:
alias (str or None): alias name or None (list-up alias names)
update (bool): force updating the alias when @alias is not None
**kwargs: keyword arguments of covsirphy.DataEngineer().subset()
[18]:
# Register
sub1, *_ = eng.subset_alias(alias="UK_with", geo="UK", complement=True)
# Retrieve with alias
sub2, *_ = eng.subset_alias(alias="UK_with")
# Comparison
sub1.equals(sub2)
[18]:
True
7. EDA of subset
With explanatory data analysis, we will get the figure of datasets.
7.1 Alias of variables
We can specify variables with alias. For example, “CIFR” is equivalent to list ['Confirmed', 'Infected', 'Recovered', 'Fatal'].
[19]:
eng.subset(geo="Japan", variables="CIRF")[0].tail()
[19]:
| Confirmed | Infected | Recovered | Fatal | |
|---|---|---|---|---|
| Date | ||||
| 2023-05-04 | 33791091 | 183209 | 33533260 | 74622 |
| 2023-05-05 | 33796902 | 177397 | 33544864 | 74641 |
| 2023-05-06 | 33803136 | 171546 | 33556937 | 74653 |
| 2023-05-07 | 33817576 | 175456 | 33567458 | 74662 |
| 2023-05-08 | 33826903 | 174762 | 33577464 | 74677 |
All aliases can be checked with DataEngineer().variables_alias().
[20]:
eng.variables_alias()
[20]:
{'N': ['Population'],
'S': ['Susceptible'],
'T': ['Tests'],
'C': ['Confirmed'],
'I': ['Infected'],
'F': ['Fatal'],
'R': ['Recovered'],
'CFR': ['Confirmed', 'Fatal', 'Recovered'],
'CIRF': ['Confirmed', 'Infected', 'Recovered', 'Fatal'],
'SIRF': ['Susceptible', 'Infected', 'Recovered', 'Fatal'],
'CR': ['Confirmed', 'Recovered']}
We can register new alias “p” with ["Tests", "Confirmed", "Positive_rate_%"] as an example.
[21]:
# Register new alias
eng.variables_alias(alias="p", variables=["Tests", "Confirmed", "Positive_rate_%"])
# Check the contents of an alias
eng.variables_alias(alias="p")
# Subsetting with the variable alias
eng.subset_alias(alias="jp", geo="Japan", variables="p")[0].tail()
[21]:
| Tests | Confirmed | Positive_rate_% | |
|---|---|---|---|
| Date | |||
| 2023-05-04 | 0.0 | 33791091 | inf |
| 2023-05-05 | 0.0 | 33796902 | inf |
| 2023-05-06 | 0.0 | 33803136 | inf |
| 2023-05-07 | 0.0 | 33817576 | inf |
| 2023-05-08 | 0.0 | 33826903 | inf |
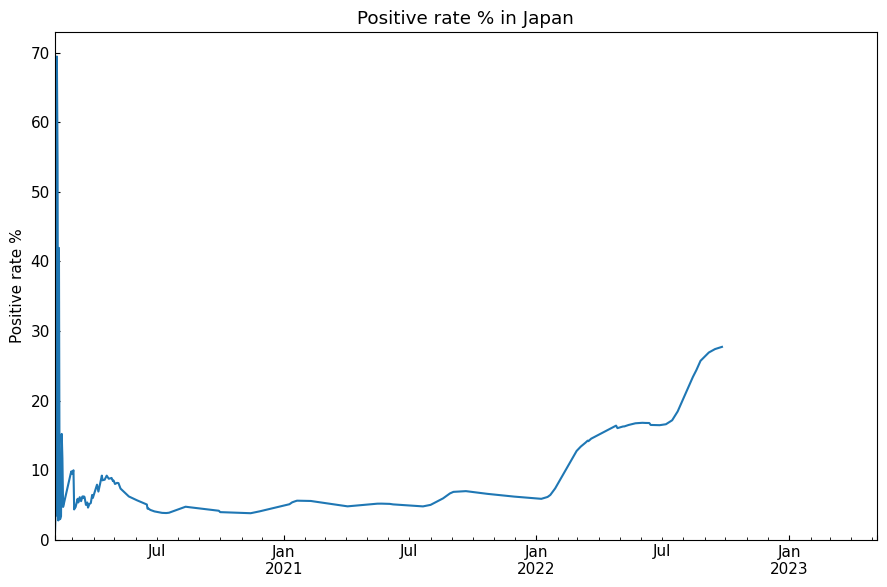
7.2 Line plot
Show data with line plot. We can select function line_plot or class LinePlot.
[22]:
line_df, *_ = eng.subset_alias(alias="jp")
With function:
[23]:
cs.line_plot(
line_df["Positive_rate_%"],
title="Positive rate % in Japan",
ylabel="Positive rate %",
math_scale=False,
show_legend=False,
filename=None,
)
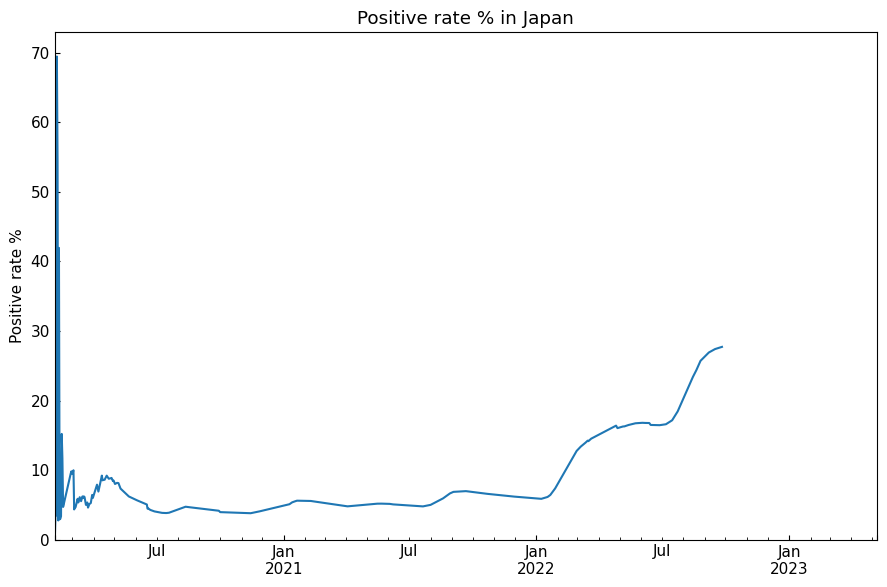
With class:
[24]:
with cs.LinePlot(filename=None) as lp:
lp.plot(line_df["Positive_rate_%"])
lp.title = "Positive rate % in Japan"
lp.x_axis(xlabel=None)
lp.y_axis(ylabel="Positive rate %", math_scale=False)
lp.legend_hide()
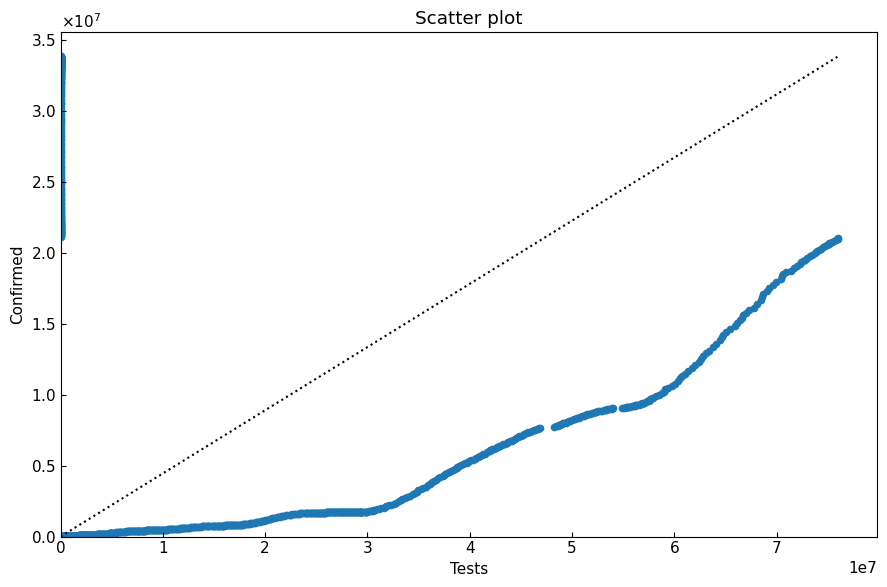
7.3 Scatter plot
Show data with scatter plot. We can select function scatter_plot or class ScatterPlot.
[25]:
sc_df, *_ = eng.subset_alias(alias="jp")
sc_df.rename(columns={"Tests": "x", "Confirmed": "y"}, inplace=True)
[26]:
cs.scatter_plot(
sc_df,
title="Scatter plot",
xlabel="Tests", xlim=(0, None),
ylabel="Confirmed",
filename=None,
)
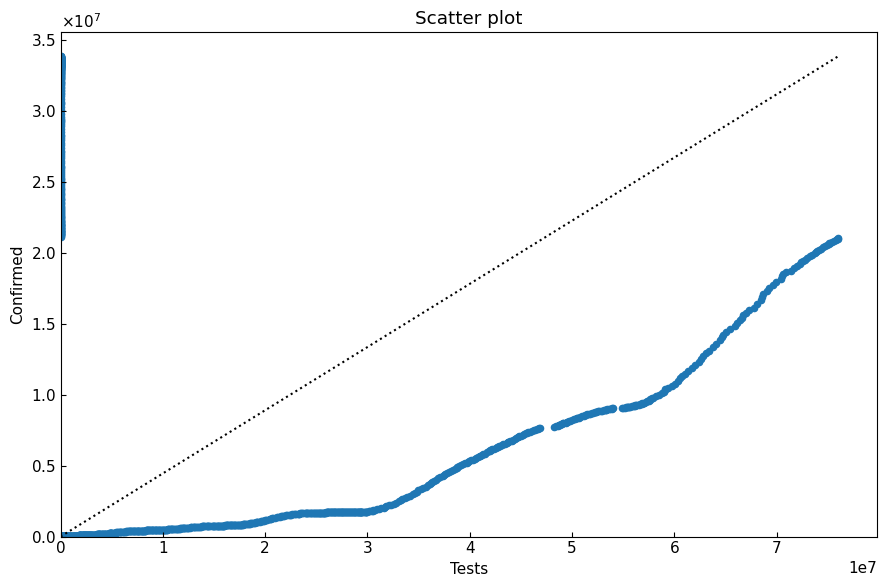
[27]:
with cs.ScatterPlot(filename=None) as sp:
sp.plot(sc_df)
sp.title = "Scatter plot"
sp.x_axis(xlabel="Tests", xlim=(0, None))
sp.y_axis(ylabel="Confirmed")
sp.line_straight(p1=(0, 0), p2=(max(sc_df["x"]), max(sc_df["y"])))
Thank you!