Data preparation
The first step of data science is data preparation. covsirphy has the following three functionality for that.
Downloading datasets from recommended data servers
Reading
pandas.DataFrameGenerator of sample data with SIR-derived ODE model
[1]:
from datetime import date
from pprint import pprint
import numpy as np
import pandas as pd
import covsirphy as cs
cs.__version__
[1]:
'3.1.2.delta'
From version 2.28.0, some classes and methods output the following information using loguru: library which aims to bring enjoyable logging in Python. We can change logging level with cs.config(level=2) (as default).
level=0: errors (exceptions)level=1: errors, warningslevel=2: errors, warnings, info (start downloading/optimization)level=3: errors, warnings, info, debug
[2]:
cs.config.logger(level=2)
1. Downloading datasets from recommended data
We will download datasets from the following recommended data servers.
-
Guidotti, E., Ardia, D., (2020), “COVID-19 Data Hub”, Journal of Open Source Software 5(51):2376, doi: 10.21105/joss.02376.
The number of cases (JHU style)
Population values in each country/province
The number of tests
-
Hasell, J., Mathieu, E., Beltekian, D. et al. A cross-country database of COVID-19 testing. Sci Data 7, 345 (2020). https://doi.org/10.1038/s41597-020-00688-8
The number of tests
The number of vaccinations
The number of people who received vaccinations
COVID-19 Open Data by Google Cloud Platform
Wahltinez and others (2020), COVID-19 Open-Data: curating a fine-grained, global-scale data repository for SARS-CoV-2, Work in progress, https://goo.gle/covid-19-open-data
percentage to baseline in visits
Note: Please refer to Google Terms of Service in advance.
This will be removed because not updated. Refer to https://github.com/lisphilar/covid19-sir/issues/1224
World Population Prospects 2022
United Nations, Department of Economic and Social Affairs, Population Division (2022). World Population Prospects 2022, Online Edition.
Total population in each country
-
Hirokazu Takaya (2020-2022), GitHub repository, COVID-19 dataset in Japan, https://github.com/lisphilar/covid19-sir/tree/master/data
The number of cases in Japan (total/prefectures)
Metadata regarding Japan prefectures
1-1. With DataEngineer class
We can use DataEngineer().download() for data downloading from recommended data servers as the quickest way.
[3]:
eng = cs.DataEngineer()
eng.download(databases=["japan", "covid19dh", "owid"])
2025-12-15 at 13:30:18 | INFO | Retrieving COVID-19 dataset from https://github.com/lisphilar/covid19-sir/data/
2025-12-15 at 13:30:18 | INFO | Retrieving datasets from COVID-19 Data Hub https://covid19datahub.io/
2025-12-15 at 13:30:23 | INFO | Retrieving datasets from Our World In Data https://github.com/owid/covid-19-data/
2025-12-15 at 13:30:24 | INFO | Retrieving datasets from Our World In Data https://github.com/owid/covid-19-data/
2025-12-15 at 13:30:25 | INFO | Retrieving datasets from Our World In Data https://github.com/owid/covid-19-data/
[3]:
<covsirphy.engineering.engineer.DataEngineer at 0x7fd7c9450ef0>
We can get the all downloaded records as a pandas.DataFrame with DataEngineer().all() method.
[4]:
all_df = eng.all()
# Overview of the records
all_df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 298052 entries, 0 to 298051
Data columns (total 27 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ISO3 298052 non-null category
1 Province 298052 non-null category
2 City 298052 non-null category
3 Date 298052 non-null datetime64[ns]
4 Cancel_events 197839 non-null Float64
5 Confirmed 239411 non-null Float64
6 Contact_tracing 197865 non-null Float64
7 Country 287385 non-null string
8 Fatal 222214 non-null Float64
9 Gatherings_restrictions 197839 non-null Float64
10 Information_campaigns 197865 non-null Float64
11 Internal_movement_restrictions 197865 non-null Float64
12 International_movement_restrictions 197872 non-null Float64
13 Population 286223 non-null Float64
14 Product 176264 non-null string
15 Recovered 74399 non-null Float64
16 School_closing 197864 non-null Float64
17 Stay_home_restrictions 197833 non-null Float64
18 Stringency_index 197828 non-null Float64
19 Testing_policy 197865 non-null Float64
20 Tests 91191 non-null Float64
21 Transport_closing 197845 non-null Float64
22 Vaccinated_full 59857 non-null Float64
23 Vaccinated_once 62933 non-null Float64
24 Vaccinations 66925 non-null Float64
25 Vaccinations_boosters 38050 non-null Float64
26 Workplace_closing 197864 non-null Float64
dtypes: Float64(21), category(3), datetime64[ns](1), string(2)
memory usage: 61.7 MB
DataEngineer.citations() shows citations of the datasets.
[5]:
print("\n".join(eng.citations()))
Hirokazu Takaya (2020-2024), COVID-19 dataset in Japan, GitHub repository, https://github.com/lisphilar/covid19-sir/data/japan
Guidotti, E., Ardia, D., (2020), "COVID-19 Data Hub", Journal of Open Source Software 5(51):2376, doi: 10.21105/joss.02376.
Hasell, J., Mathieu, E., Beltekian, D. et al. A cross-country database of COVID-19 testing. Sci Data 7, 345 (2020). https: //doi.org/10.1038/s41597-020-00688-8
Note that, as default, DataEngineer().download() collects country-level data and save the datasets as CSV files in “input” (=directory argument of DataEngineer().download()) folder of the current directory. If the last modification time of the saved CSV files is within the last 12 (=update_interval argument of DataEngineer().download()) hours, the saved CSV files will be used as the database.
For some countries (eg. Japan), province/state/prefecture level data is available and we can download it as follows.
[6]:
eng_jpn = cs.DataEngineer()
eng_jpn.download(country="Japan", databases=["japan", "covid19dh", "owid"])
eng_jpn.all().head()
2025-12-15 at 13:30:27 | INFO | Retrieving COVID-19 dataset from https://github.com/lisphilar/covid19-sir/data/
2025-12-15 at 13:30:28 | INFO | Retrieving datasets from COVID-19 Data Hub https://covid19datahub.io/
[6]:
| ISO3 | Province | City | Date | Cancel_events | Confirmed | Contact_tracing | Country | Fatal | Gatherings_restrictions | ... | Stay_home_restrictions | Stringency_index | Testing_policy | Tests | Transport_closing | Vaccinated_full | Vaccinated_once | Vaccinations | Vaccinations_boosters | Workplace_closing | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | JPN | Aichi | - | 2020-03-18 | 1.0 | 130.0 | 2.0 | Japan | 14.0 | 0.0 | ... | 0.0 | -40.74 | 1.0 | 847.0 | 0.0 | None | None | None | None | 1.0 |
| 1 | JPN | Aichi | - | 2020-03-19 | 1.0 | 134.0 | 2.0 | Japan | 14.0 | 0.0 | ... | 0.0 | -40.74 | 1.0 | 847.0 | 0.0 | None | None | None | None | 1.0 |
| 2 | JPN | Aichi | - | 2020-03-20 | 1.0 | 139.0 | 2.0 | Japan | 16.0 | 0.0 | ... | 0.0 | -40.74 | 1.0 | 847.0 | 0.0 | None | None | None | None | 1.0 |
| 3 | JPN | Aichi | - | 2020-03-21 | 1.0 | 141.0 | 2.0 | Japan | 16.0 | 0.0 | ... | 0.0 | -40.74 | 1.0 | 847.0 | 0.0 | None | None | None | None | 1.0 |
| 4 | JPN | Aichi | - | 2020-03-22 | 1.0 | 143.0 | 2.0 | Japan | 16.0 | 0.0 | ... | 0.0 | -40.74 | 1.0 | 847.0 | 0.0 | None | None | None | None | 1.0 |
5 rows × 26 columns
For some countries (eg. USA), city-level data is available and we can download it as follows.
[7]:
eng_alabama = cs.DataEngineer()
eng_alabama.download(country="USA", province="Alabama", databases=["japan", "covid19dh", "owid"])
eng_alabama.all().head()
2025-12-15 at 13:30:29 | INFO | Retrieving datasets from COVID-19 Data Hub https://covid19datahub.io/
[7]:
| ISO3 | Province | City | Date | Cancel_events | Confirmed | Contact_tracing | Country | Fatal | Gatherings_restrictions | ... | International_movement_restrictions | Population | Recovered | School_closing | Stay_home_restrictions | Stringency_index | Testing_policy | Tests | Transport_closing | Workplace_closing | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | USA | Alabama | Autauga | 2020-03-24 | -2.0 | 1.0 | 1.0 | United States | 0.0 | -4.0 | ... | 3.0 | 55869.0 | <NA> | 3.0 | -2.0 | -68.98 | 1.0 | <NA> | -1.0 | -3.0 |
| 1 | USA | Alabama | Autauga | 2020-03-25 | -2.0 | 4.0 | 1.0 | United States | 0.0 | -4.0 | ... | 3.0 | 55869.0 | <NA> | 3.0 | -2.0 | -68.98 | 1.0 | <NA> | -1.0 | -3.0 |
| 2 | USA | Alabama | Autauga | 2020-03-26 | -2.0 | 6.0 | 1.0 | United States | 0.0 | -4.0 | ... | 3.0 | 55869.0 | <NA> | 3.0 | -2.0 | -68.98 | 1.0 | <NA> | -1.0 | -3.0 |
| 3 | USA | Alabama | Autauga | 2020-03-27 | -2.0 | 6.0 | 1.0 | United States | 0.0 | -4.0 | ... | 3.0 | 55869.0 | <NA> | 3.0 | -2.0 | -68.98 | 1.0 | <NA> | -1.0 | -3.0 |
| 4 | USA | Alabama | Autauga | 2020-03-28 | 2.0 | 6.0 | 1.0 | United States | 0.0 | -4.0 | ... | 3.0 | 55869.0 | <NA> | 3.0 | -2.0 | -73.61 | 1.0 | <NA> | -1.0 | 3.0 |
5 rows × 22 columns
Move forward to Tutorial: Data engineering.
1-2. With DataDownloader class
DataEngineer class is useful because it has data cleaning methods and so on (explained with Tutorial: Data engineering), but we can use DataDownloader class for data downloading.
[8]:
dl = cs.DataDownloader()
dl_df = dl.layer(country=None, province=None, databases=["japan", "covid19dh", "owid"])
[9]:
# Overview of the records
dl_df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 298052 entries, 0 to 298051
Data columns (total 27 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ISO3 298052 non-null string
1 Province 298052 non-null string
2 City 298052 non-null string
3 Date 298052 non-null datetime64[ns]
4 Cancel_events 197839 non-null Int64
5 Confirmed 239411 non-null Float64
6 Contact_tracing 197865 non-null Int64
7 Country 287385 non-null string
8 Fatal 222214 non-null Float64
9 Gatherings_restrictions 197839 non-null Int64
10 Information_campaigns 197865 non-null Int64
11 Internal_movement_restrictions 197865 non-null Int64
12 International_movement_restrictions 197872 non-null Int64
13 Population 286223 non-null Int64
14 Product 176264 non-null string
15 Recovered 74399 non-null Float64
16 School_closing 197864 non-null Int64
17 Stay_home_restrictions 197833 non-null Int64
18 Stringency_index 197828 non-null Float64
19 Testing_policy 197865 non-null Int64
20 Tests 91191 non-null Float64
21 Transport_closing 197845 non-null Int64
22 Vaccinated_full 59857 non-null Float64
23 Vaccinated_once 62933 non-null Float64
24 Vaccinations 66925 non-null Float64
25 Vaccinations_boosters 38050 non-null Float64
26 Workplace_closing 197864 non-null Int64
dtypes: Float64(9), Int64(12), datetime64[ns](1), string(5)
memory usage: 67.4 MB
Note that ISO3/Province/City columns have string data instead of categorical data.
[10]:
# Citations
print("\n".join(dl.citations()))
Hirokazu Takaya (2020-2024), COVID-19 dataset in Japan, GitHub repository, https://github.com/lisphilar/covid19-sir/data/japan
Guidotti, E., Ardia, D., (2020), "COVID-19 Data Hub", Journal of Open Source Software 5(51):2376, doi: 10.21105/joss.02376.
Hasell, J., Mathieu, E., Beltekian, D. et al. A cross-country database of COVID-19 testing. Sci Data 7, 345 (2020). https: //doi.org/10.1038/s41597-020-00688-8
Acknowledgement
In Feb2020, CovsirPhy project started in Kaggle platform with COVID-19 data with SIR model notebook by Hirokazu Takaya helped by Kagglers using the following datasets.
The number of cases (JHU) and linelist: Novel Corona Virus 2019 Dataset by SRK
Population in each country: covid19 global forecasting: locations population by Dmitry A. Grechka
The number of cases in Japan: COVID-19 dataset in Japan by Lisphilar
The current version of covsirphy does not have interfaces to use the datasets in Kaggle because they are not updated at this time. However, we could not have done CovsirPhy project without their supports. Thank you!!
2. Reading pandas.DataFrame
We may need to use our own datasets for analysis because the dataset is not included in the recommended data servers. DataEngineer().register() registers new datasets of pandas.DataFrame format.
2-1. Retrieve Monkeypox line list
At first, we will prepare the new dataset as pandas.DataFrame. We will use Global.health Monkeypox under CC BY 4.0 license, line list data regarding Monkeypox 2022, just for demonstration.
[11]:
today = date.today()
mp_cite = f"Global.health Monkeypox (accessed on {today.strftime('%Y-%m-%d')}):\n" \
"Kraemer, Tegally, Pigott, Dasgupta, Sheldon, Wilkinson, Schultheiss, et al. " \
"Tracking the 2022 Monkeypox Outbreak with Epidemiological Data in Real-Time. " \
"The Lancet Infectious Diseases. https://doi.org/10.1016/S1473-3099(22)00359-0.\n" \
"European Centre for Disease Prevention and Control/WHO Regional Office for Europe." \
f" Monkeypox, Joint Epidemiological overview, {today.day} {today.month}, 2022"
print(mp_cite)
Global.health Monkeypox (accessed on 2025-12-15):
Kraemer, Tegally, Pigott, Dasgupta, Sheldon, Wilkinson, Schultheiss, et al. Tracking the 2022 Monkeypox Outbreak with Epidemiological Data in Real-Time. The Lancet Infectious Diseases. https://doi.org/10.1016/S1473-3099(22)00359-0.
European Centre for Disease Prevention and Control/WHO Regional Office for Europe. Monkeypox, Joint Epidemiological overview, 15 12, 2022
Retrieve CSV file with pandas.read_csv(), using Pyarrow as the engine.
[12]:
# Deprecated: Final line list from Global.health, as of 2022-09-22
raw_url = "https://raw.githubusercontent.com/globaldothealth/monkeypox/9035dfb1303c05c380f86a3c1e00f81a1cc4046b/latest_deprecated.csv"
raw = pd.read_csv(raw_url, engine="pyarrow")
raw.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 69595 entries, 0 to 69594
Data columns (total 36 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ID 69595 non-null object
1 Status 69595 non-null object
2 Location 52387 non-null object
3 City 1383 non-null object
4 Country 69595 non-null object
5 Country_ISO3 69595 non-null object
6 Age 3021 non-null object
7 Gender 2475 non-null object
8 Date_onset 77 non-null object
9 Date_confirmation 65546 non-null object
10 Symptoms 220 non-null object
11 Hospitalised (Y/N/NA) 354 non-null object
12 Date_hospitalisation 35 non-null object
13 Isolated (Y/N/NA) 493 non-null object
14 Date_isolation 16 non-null object
15 Outcome 101 non-null object
16 Contact_comment 91 non-null object
17 Contact_ID 27 non-null float64
18 Contact_location 6 non-null object
19 Travel_history (Y/N/NA) 365 non-null object
20 Travel_history_entry 42 non-null object
21 Travel_history_start 11 non-null object
22 Travel_history_location 114 non-null object
23 Travel_history_country 99 non-null object
24 Genomics_Metadata 24 non-null object
25 Confirmation_method 100 non-null object
26 Source 69595 non-null object
27 Source_II 8240 non-null object
28 Source_III 893 non-null object
29 Source_IV 54 non-null object
30 Source_V 0 non-null float64
31 Source_VI 0 non-null float64
32 Source_VII 0 non-null float64
33 Date_entry 69595 non-null object
34 Date_death 83 non-null object
35 Date_last_modified 69595 non-null object
dtypes: float64(4), object(32)
memory usage: 19.1+ MB
Review the data.
[13]:
raw.head()
[13]:
| ID | Status | Location | City | Country | Country_ISO3 | Age | Gender | Date_onset | Date_confirmation | ... | Source | Source_II | Source_III | Source_IV | Source_V | Source_VI | Source_VII | Date_entry | Date_death | Date_last_modified | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | N1 | confirmed | Guy's and St Thomas Hospital London | London | England | GBR | None | None | 2022-04-29 | 2022-05-06 | ... | https://www.gov.uk/government/news/monkeypox-c... | https://www.who.int/emergencies/disease-outbre... | None | None | NaN | NaN | NaN | 2022-05-18 | None | 2022-05-18 |
| 1 | N2 | confirmed | Guy's and St Thomas Hospital London | London | England | GBR | None | None | 2022-05-05 | 2022-05-12 | ... | https://www.gov.uk/government/news/monkeypox-c... | None | None | None | NaN | NaN | NaN | 2022-05-18 | None | 2022-05-18 |
| 2 | N3 | confirmed | London | London | England | GBR | None | None | 2022-04-30 | 2022-05-13 | ... | https://www.gov.uk/government/news/monkeypox-c... | None | None | None | NaN | NaN | NaN | 2022-05-18 | None | 2022-05-18 |
| 3 | N4 | confirmed | London | London | England | GBR | None | male | None | 2022-05-15 | ... | https://www.gov.uk/government/news/monkeypox-c... | None | None | None | NaN | NaN | NaN | 2022-05-18 | None | 2022-05-18 |
| 4 | N5 | confirmed | London | London | England | GBR | None | male | None | 2022-05-15 | ... | https://www.gov.uk/government/news/monkeypox-c... | None | None | None | NaN | NaN | NaN | 2022-05-18 | None | 2022-05-18 |
5 rows × 36 columns
[14]:
pprint(raw.Status.unique())
pprint(raw.Outcome.unique())
array(['confirmed', 'discarded', 'suspected', 'omit_error'], dtype=object)
array([None, 'Recovered', 'Death'], dtype=object)
2-2. Convert line list to the number of cases data
Prepare analyzable data, converting the line list to the number of cases. This step may be skipped because we have datasets with the number of cases.
Prepare PPT (per protocol set) data.
[15]:
date_cols = [
"Date_onset", "Date_confirmation", "Date_hospitalisation",
"Date_isolation", "Date_death", "Date_last_modified"
]
cols = ["ID", "Status", "City", "Country_ISO3", "Outcome", *date_cols]
df = raw.loc[:, cols].rename(columns={"Country_ISO3": "ISO3"})
df = df.loc[df["Status"].isin(["confirmed", "suspected"])]
for col in date_cols:
df[col] = pd.to_datetime(df[col])
df["Date_min"] = df[date_cols].min(axis=1)
df["Date_recovered"] = df[["Outcome", "Date_last_modified"]].apply(
lambda x: x[1] if x[0] == "Recovered" else pd.NaT, axis=1)
df["City"] = df["City"].replace('', pd.NA).fillna("Unknown")
ppt_df = df.copy()
ppt_df.head()
[15]:
| ID | Status | City | ISO3 | Outcome | Date_onset | Date_confirmation | Date_hospitalisation | Date_isolation | Date_death | Date_last_modified | Date_min | Date_recovered | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | N1 | confirmed | London | GBR | None | 2022-04-29 | 2022-05-06 | 2022-05-04 | 2022-05-04 | NaT | 2022-05-18 | 2022-04-29 | NaT |
| 1 | N2 | confirmed | London | GBR | None | 2022-05-05 | 2022-05-12 | 2022-05-06 | 2022-05-09 | NaT | 2022-05-18 | 2022-05-05 | NaT |
| 2 | N3 | confirmed | London | GBR | None | 2022-04-30 | 2022-05-13 | NaT | NaT | NaT | 2022-05-18 | 2022-04-30 | NaT |
| 3 | N4 | confirmed | London | GBR | None | NaT | 2022-05-15 | NaT | NaT | NaT | 2022-05-18 | 2022-05-15 | NaT |
| 4 | N5 | confirmed | London | GBR | None | NaT | 2022-05-15 | NaT | NaT | NaT | 2022-05-18 | 2022-05-15 | NaT |
Calculate daily new confirmed cases.
[16]:
df = ppt_df.rename(columns={"Date_min": "Date"})
series = df.groupby(["ISO3", "City", "Date"])["ID"].count()
series.name = "Confirmed"
c_df = pd.DataFrame(series)
c_df.head()
[16]:
| Confirmed | |||
|---|---|---|---|
| ISO3 | City | Date | |
| ABW | Unknown | 2022-08-22 | 1 |
| 2022-08-29 | 1 | ||
| 2022-09-13 | 1 | ||
| AND | Unknown | 2022-07-25 | 1 |
| 2022-07-26 | 2 |
Calculate daily new recovered cases.
[17]:
df = ppt_df.rename(columns={"Date_recovered": "Date"})
series = df.groupby(["ISO3", "City", "Date"])["ID"].count()
series.name = "Recovered"
r_df = pd.DataFrame(series)
r_df.head()
[17]:
| Recovered | |||
|---|---|---|---|
| ISO3 | City | Date | |
| AUS | Melbourne | 2022-06-30 | 1 |
| TUR | Unknown | 2022-08-05 | 4 |
Calculate daily new fatal cases.
[18]:
df = ppt_df.rename(columns={"Date_death": "Date"})
series = df.groupby(["ISO3", "City", "Date"])["ID"].count()
series.name = "Fatal"
f_df = pd.DataFrame(series)
f_df.head()
[18]:
| Fatal | |||
|---|---|---|---|
| ISO3 | City | Date | |
| BEL | Unknown | 2022-08-29 | 1 |
| BRA | Unknown | 2022-07-28 | 1 |
| 2022-08-29 | 1 | ||
| CAF | Unknown | 2022-03-04 | 2 |
| COD | Unknown | 2022-01-16 | 18 |
Combine data (cumulative number).
[19]:
df = c_df.combine_first(f_df).combine_first(r_df)
df = df.unstack(level=["ISO3", "City"])
df = df.asfreq("D").fillna(0).cumsum()
df = df.stack(level=["ISO3", "City"]).reorder_levels(["ISO3", "City", "Date"])
df = df.sort_index().reset_index()
all_df_city = df.copy()
all_df_city.head()
[19]:
| ISO3 | City | Date | Confirmed | Fatal | Recovered | |
|---|---|---|---|---|---|---|
| 0 | ABW | Unknown | 2022-01-16 | 0.0 | 0.0 | 0.0 |
| 1 | ABW | Unknown | 2022-01-17 | 0.0 | 0.0 | 0.0 |
| 2 | ABW | Unknown | 2022-01-18 | 0.0 | 0.0 | 0.0 |
| 3 | ABW | Unknown | 2022-01-19 | 0.0 | 0.0 | 0.0 |
| 4 | ABW | Unknown | 2022-01-20 | 0.0 | 0.0 | 0.0 |
At country level (City = “-”) and city level (City != “-“):
[20]:
df2 = all_df_city.groupby(["ISO3", "Date"], as_index=False).sum()
df2["City"] = "-"
df = pd.concat([df2, all_df_city], axis=0)
df = df.loc[df["City"] != "Unknown"]
all_df = df.convert_dtypes()
all_df
[20]:
| ISO3 | Date | City | Confirmed | Fatal | Recovered | |
|---|---|---|---|---|---|---|
| 0 | ABW | 2022-01-16 | - | 0 | 0 | 0 |
| 1 | ABW | 2022-01-17 | - | 0 | 0 | 0 |
| 2 | ABW | 2022-01-18 | - | 0 | 0 | 0 |
| 3 | ABW | 2022-01-19 | - | 0 | 0 | 0 |
| 4 | ABW | 2022-01-20 | - | 0 | 0 | 0 |
| ... | ... | ... | ... | ... | ... | ... |
| 69245 | ZAF | 2022-09-18 | Johannesburg | 1 | 0 | 0 |
| 69246 | ZAF | 2022-09-19 | Johannesburg | 1 | 0 | 0 |
| 69247 | ZAF | 2022-09-20 | Johannesburg | 1 | 0 | 0 |
| 69248 | ZAF | 2022-09-21 | Johannesburg | 1 | 0 | 0 |
| 69249 | ZAF | 2022-09-22 | Johannesburg | 1 | 0 | 0 |
70500 rows × 6 columns
Check data.
[21]:
gis = cs.GIS(layers=["ISO3", "City"], country="ISO3", date="Date")
gis.register(data=all_df, convert_iso3=False);
[22]:
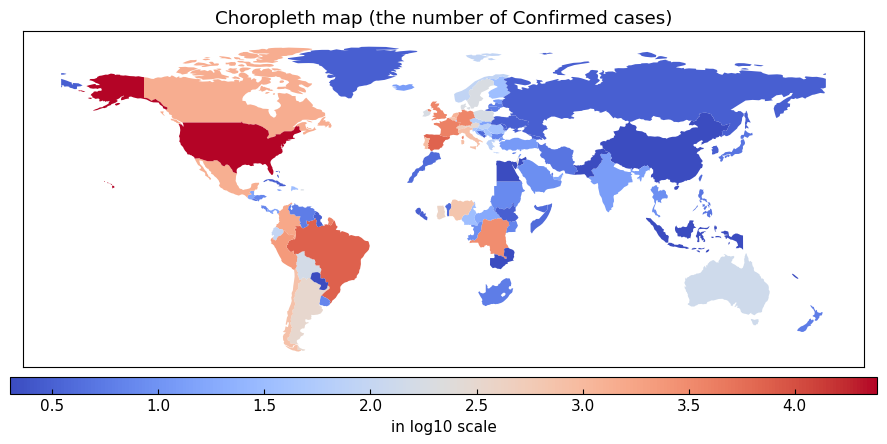
variable = "Confirmed"
gis.choropleth(
variable=variable, filename=None,
title=f"Choropleth map (the number of {variable} cases)"
)
2025-12-15 at 13:31:17 | INFO | Retrieving GIS data from Natural Earth https://www.naturalearthdata.com/
[23]:
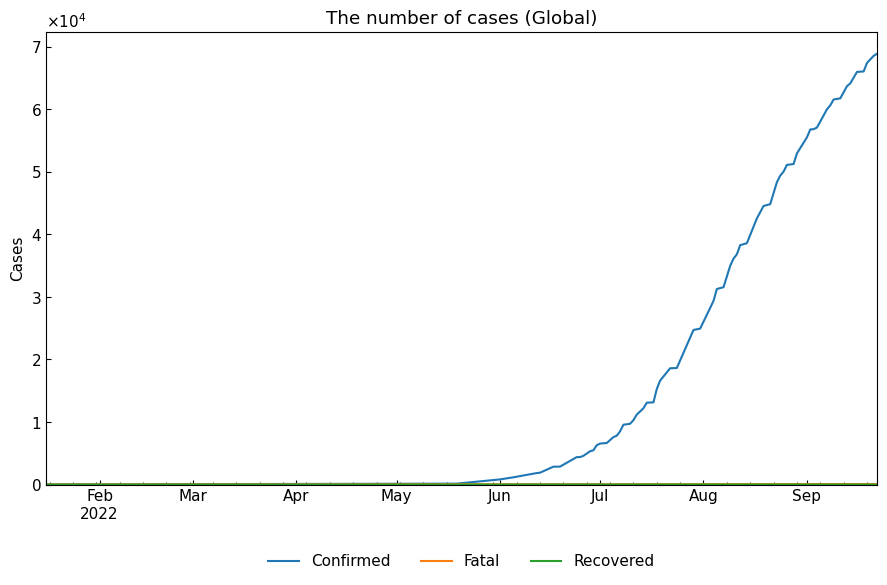
global_df = gis.subset(geo=None).set_index("Date").astype(np.int64)
global_df.tail()
cs.line_plot(global_df, title="The number of cases (Global)")
2-3. Retrieve total population data
So that we can analyze the data, total population values are necessary (we will confirm this with Tutorial: SIR-derived ODE models later).
Population data at country-level can be retrieved with DataDownloader().layer(databases=["wpp"]) via DataEngineer().register(databases=["wpp"]).
[24]:
# Set layers and specify layer name of country
# (which will be converted to ISO3 code for standardization)
eng = cs.DataEngineer(layers=["ISO3", "City"], country=["ISO3"])
# Download and automated registration of population data
eng.download(databases=["wpp"])
# Specify date range to reduce the memory
date_range = (all_df["Date"].min(), all_df["Date"].max())
eng.clean(kinds=["resample"], date_range=date_range)
# Show all data
display(eng.all())
# Show citations
pprint(eng.citations())
2025-12-15 at 13:31:20 | INFO | [INFO] 'Province' layer was removed.
| ISO3 | City | Date | Population | |
|---|---|---|---|---|
| 0 | ABW | - | 2022-07-01 | 107310.0 |
| 1 | AFE | - | 2022-07-01 | 731821393.0 |
| 2 | AFG | - | 2022-07-01 | 40578842.0 |
| 3 | AFW | - | 2022-07-01 | 497387180.0 |
| 4 | AGO | - | 2022-07-01 | 35635029.0 |
| ... | ... | ... | ... | ... |
| 256 | XKX | - | 2022-07-01 | 1768096.0 |
| 257 | YEM | - | 2022-07-01 | 38222876.0 |
| 258 | ZAF | - | 2022-07-01 | 62378410.0 |
| 259 | ZMB | - | 2022-07-01 | 20152938.0 |
| 260 | ZWE | - | 2022-07-01 | 16069056.0 |
261 rows × 4 columns
['United Nations, Department of Economic and Social Affairs, Population '
'Division (2022). World Population Prospects 2022, Online Edition.']
2-4. Register Monkeypox data
Register the Monkeypox data to DataEngineer() instance.
[25]:
eng.register(data=all_df, citations=[mp_cite])
# Show all data
display(eng.all())
# Show citations
pprint(eng.citations())
| ISO3 | City | Date | Confirmed | Fatal | Population | Recovered | |
|---|---|---|---|---|---|---|---|
| 0 | ABW | - | 2022-01-16 | 0 | 0 | <NA> | 0 |
| 1 | ABW | - | 2022-01-17 | 0 | 0 | <NA> | 0 |
| 2 | ABW | - | 2022-01-18 | 0 | 0 | <NA> | 0 |
| 3 | ABW | - | 2022-01-19 | 0 | 0 | <NA> | 0 |
| 4 | ABW | - | 2022-01-20 | 0 | 0 | <NA> | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 70644 | ZMB | - | 2022-09-19 | 1 | 0 | <NA> | 0 |
| 70645 | ZMB | - | 2022-09-20 | 1 | 0 | <NA> | 0 |
| 70646 | ZMB | - | 2022-09-21 | 1 | 0 | <NA> | 0 |
| 70647 | ZMB | - | 2022-09-22 | 1 | 0 | <NA> | 0 |
| 70648 | ZWE | - | 2022-07-01 | <NA> | <NA> | 16069056.0 | <NA> |
70649 rows × 7 columns
['United Nations, Department of Economic and Social Affairs, Population '
'Division (2022). World Population Prospects 2022, Online Edition.',
'Global.health Monkeypox (accessed on 2025-12-15):\n'
'Kraemer, Tegally, Pigott, Dasgupta, Sheldon, Wilkinson, Schultheiss, et al. '
'Tracking the 2022 Monkeypox Outbreak with Epidemiological Data in Real-Time. '
'The Lancet Infectious Diseases. '
'https://doi.org/10.1016/S1473-3099(22)00359-0.\n'
'European Centre for Disease Prevention and Control/WHO Regional Office for '
'Europe. Monkeypox, Joint Epidemiological overview, 15 12, 2022']
Move forward to Tutorial: Data engineering.
3. Generator of sample data with SIR-derived ODE model
CovsirPhy can generate sample data with subclasses of ODEModel and Dynamics class. Refer to the followings.
3.1 Sample data of one-phase ODE model
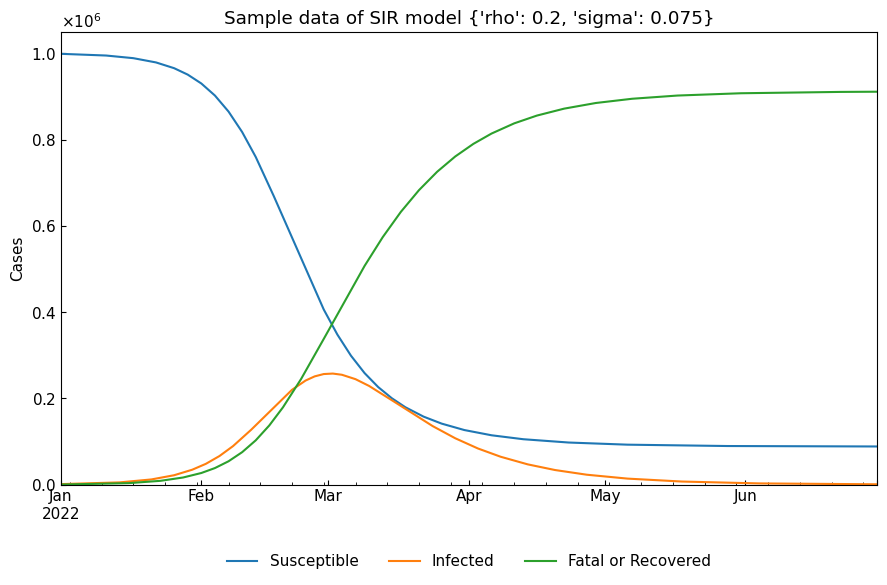
Regarding ODE models, please refer to Tutorial: SIR-derived ODE models. Here, we will create a sample data with one-phase SIR model and tau value 1440 min, the first date 01Jan2022, the last date 30Jun2022. ODE parameter values are preset.
[26]:
# Create solver with preset
model = cs.SIRModel.from_sample(date_range=("01Jan2022", "30Jun2022"), tau=1440)
# Show settings
pprint(model.settings())
{'date_range': ('01Jan2022', '30Jun2022'),
'initial_dict': {'Fatal or Recovered': 0,
'Infected': 1000,
'Susceptible': 999000},
'param_dict': {'rho': 0.2, 'sigma': 0.075},
'tau': 1440}
Solve the ODE model with ODEModel().solve() method.
[27]:
one_df = model.solve()
display(one_df.head())
display(one_df.tail())
| Susceptible | Infected | Fatal or Recovered | |
|---|---|---|---|
| Date | |||
| 2022-01-01 | 999000 | 1000 | 0 |
| 2022-01-02 | 998787 | 1133 | 80 |
| 2022-01-03 | 998546 | 1283 | 170 |
| 2022-01-04 | 998273 | 1454 | 273 |
| 2022-01-05 | 997964 | 1647 | 389 |
| Susceptible | Infected | Fatal or Recovered | |
|---|---|---|---|
| Date | |||
| 2022-06-26 | 88354 | 750 | 910895 |
| 2022-06-27 | 88342 | 708 | 910950 |
| 2022-06-28 | 88329 | 669 | 911002 |
| 2022-06-29 | 88318 | 632 | 911050 |
| 2022-06-30 | 88307 | 596 | 911096 |
Plot the time-series data.
[28]:
cs.line_plot(one_df, title=f"Sample data of SIR model {model.settings()['param_dict']}")
3.2 Sample data of multi-phase ODE model
Regarding multi-phase ODE models, please refer to Phase-dependent SIR models. Here, we will create a sample data with two-phase SIR model and tau value 1440 min, the first date 01Jan2022, the last date 30Jun2022.
We will use Dynamics class. At first, set the first/date of dynamics and set th 0th phase ODE parameters.
[29]:
dyn = cs.Dynamics.from_sample(model=cs.SIRModel, date_range=("01Jan2022", "30Jun2022"))
# Show summary
dyn.summary()
[29]:
| Start | End | Rt | rho | sigma | 1/beta [day] | 1/gamma [day] | |
|---|---|---|---|---|---|---|---|
| Phase | |||||||
| 0th | 2022-01-01 | 2022-06-30 | 2.67 | 0.2 | 0.075 | 5 | 13 |
Add the 1st phase with Dynamics.register() method.
[30]:
setting_df = dyn.register()
setting_df.loc["01Mar2022": "30Jun2022", ["rho", "sigma"]] = [0.4, 0.075]
setting_df
[30]:
| Susceptible | Infected | Recovered | Fatal | rho | sigma | |
|---|---|---|---|---|---|---|
| Date | ||||||
| 2022-01-01 | 999000.0 | 1000.0 | 0.0 | 0.0 | 0.2 | 0.075 |
| 2022-01-02 | <NA> | <NA> | <NA> | <NA> | <NA> | <NA> |
| 2022-01-03 | <NA> | <NA> | <NA> | <NA> | <NA> | <NA> |
| 2022-01-04 | <NA> | <NA> | <NA> | <NA> | <NA> | <NA> |
| 2022-01-05 | <NA> | <NA> | <NA> | <NA> | <NA> | <NA> |
| ... | ... | ... | ... | ... | ... | ... |
| 2022-06-26 | <NA> | <NA> | <NA> | <NA> | 0.4 | 0.075 |
| 2022-06-27 | <NA> | <NA> | <NA> | <NA> | 0.4 | 0.075 |
| 2022-06-28 | <NA> | <NA> | <NA> | <NA> | 0.4 | 0.075 |
| 2022-06-29 | <NA> | <NA> | <NA> | <NA> | 0.4 | 0.075 |
| 2022-06-30 | <NA> | <NA> | <NA> | <NA> | 0.4 | 0.075 |
181 rows × 6 columns
[31]:
dyn.register(data=setting_df)
# Show summary
dyn.summary()
[31]:
| Start | End | Rt | rho | sigma | 1/beta [day] | 1/gamma [day] | |
|---|---|---|---|---|---|---|---|
| Phase | |||||||
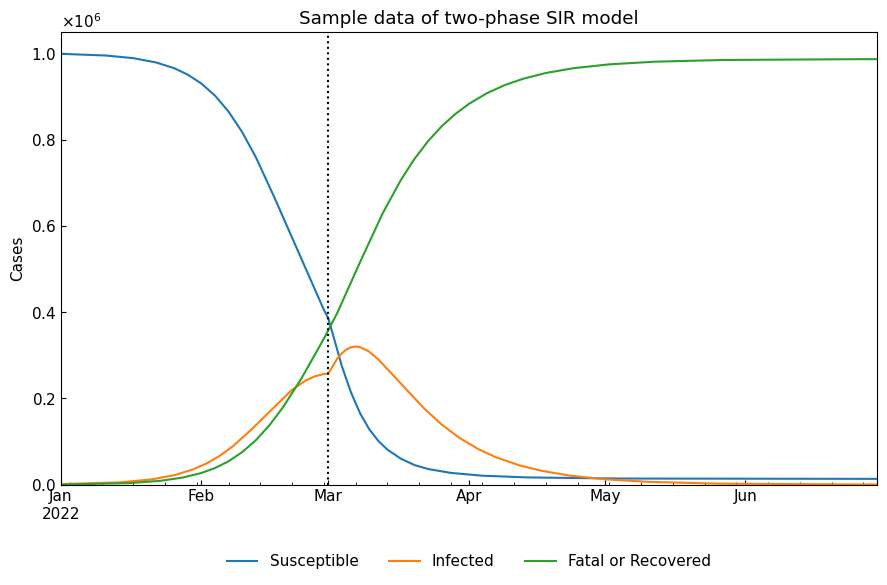
| 0th | 2022-01-01 | 2022-02-28 | 2.67 | 0.2 | 0.075 | 5 | 13 |
| 1st | 2022-03-01 | 2022-06-30 | 5.33 | 0.4 | 0.075 | 2 | 13 |
Solve the ODE model with Dynamics().simulate() method and plot the time-series data.
[32]:
two_df = dyn.simulate(model_specific=True)
cs.line_plot(two_df, title="Sample data of two-phase SIR model", v=["01Mar2022"])
When we need convert model-specific variables to model-free variables (Susceptible/Infected/Fatal/Recovered), we will set model_specific=False (default). Because R=”Fatal or Recovered” in SIR model, we assume that R=”Recovered” and F = 0.
[33]:
two_df = dyn.simulate(model_specific=False)
cs.line_plot(two_df, title="Sample data of two-phase SIR model with SIRF variables", v=["01Mar2022"])
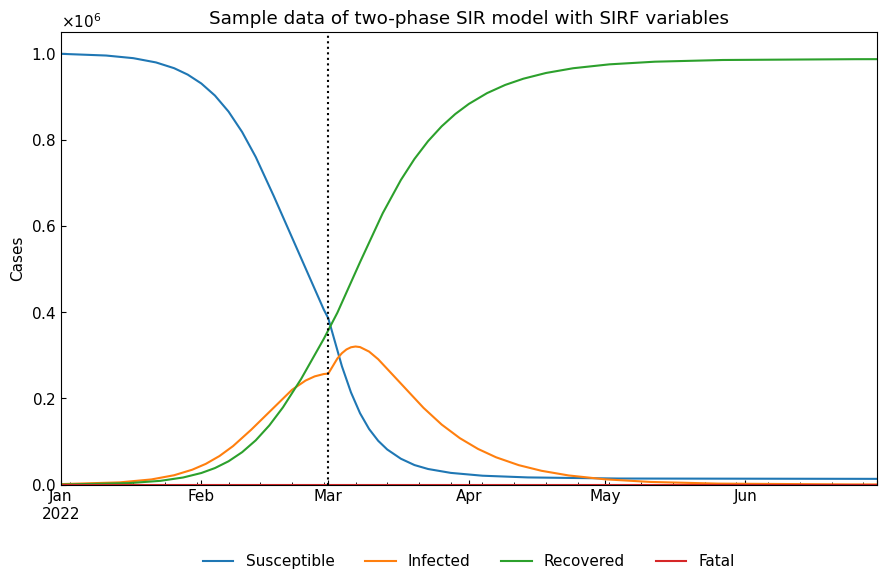
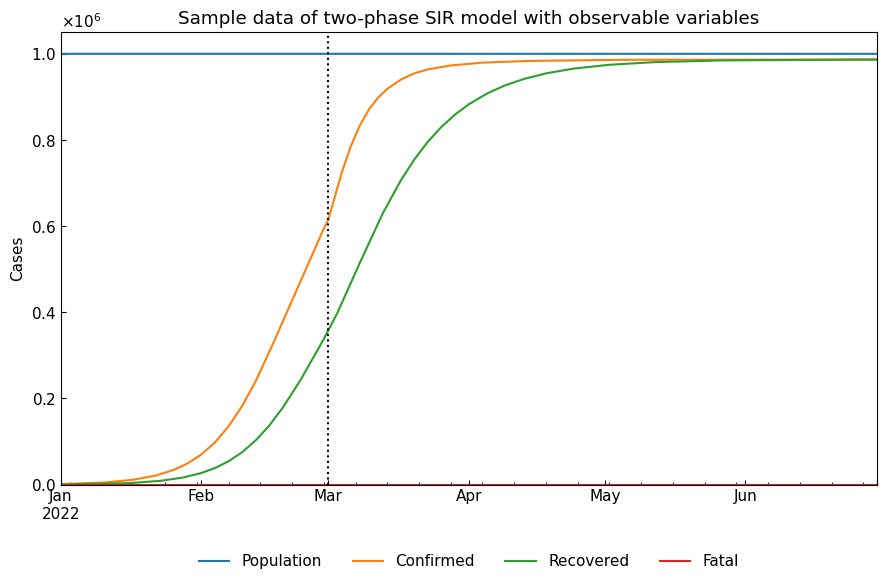
Actually, observable variables are Population/Confirmed/Infected/Recovered. We can calculate Population and Confirmed as follows.
Confirmed = Infected + Fatal + Recovered
Population = Susceptible + Confirmed
[34]:
real_df = two_df.copy()
real_df["Confirmed"] = real_df[["Infected", "Fatal", "Recovered"]].sum(axis=1)
real_df["Population"] = real_df[["Susceptible", "Confirmed"]].sum(axis=1)
real_df = real_df.loc[:, ["Population", "Confirmed", "Recovered", "Fatal"]]
cs.line_plot(real_df, title="Sample data of two-phase SIR model with observable variables", v=["01Mar2022"])
Thank you!