.. raw:: html
 |PyPI version| |Downloads| |PyPI - Python Version| |GitHub license|
|Quality Check| |Test Coverage|
CovsirPhy introduction
======================
`Documentation `__
\|
`Installation `__
\|
`Tutorial `__
\| `API
reference `__ \|
`GitHub `__ \| `Qiita
(Japanese) `__
CovsirPhy is a Python library for infectious disease (COVID-19:
Coronavirus disease 2019, Monkeypox 2022) data analysis with
phase-dependent SIR-derived ODE models. We can download datasets and
analyze them easily. Scenario analysis with CovsirPhy enables us to make
data-informed decisions.
.. raw:: html
|PyPI version| |Downloads| |PyPI - Python Version| |GitHub license|
|Quality Check| |Test Coverage|
CovsirPhy introduction
======================
`Documentation `__
\|
`Installation `__
\|
`Tutorial `__
\| `API
reference `__ \|
`GitHub `__ \| `Qiita
(Japanese) `__
CovsirPhy is a Python library for infectious disease (COVID-19:
Coronavirus disease 2019, Monkeypox 2022) data analysis with
phase-dependent SIR-derived ODE models. We can download datasets and
analyze them easily. Scenario analysis with CovsirPhy enables us to make
data-informed decisions.
.. raw:: html
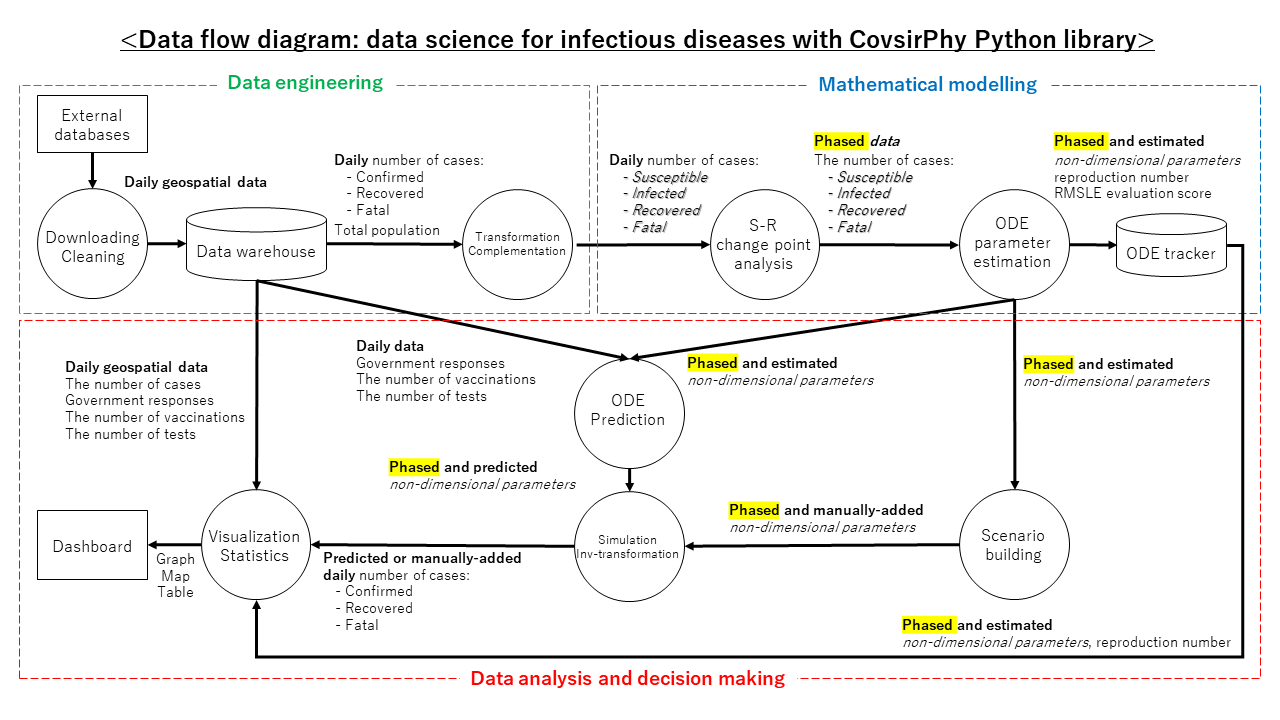 Inspiration
-----------
- Monitor the spread of COVID-19/Monkeypox with SIR-derived ODE models
- Predict the number of cases in each country/province
- Find the relationship of reproductive number and measures taken by
each country
If you have ideas or need new functionalities, please join this project.
Any suggestions with `Github
Issues `__
and `Twitter: @lisphilar `__ are always
welcomed. Questions are also great. Please refer to `Guideline of
contribution `__.
Installation
------------
The latest stable version of CovsirPhy is available at `PyPI (The Python
Package Index): covsirphy `__ and
supports Python 3.8 or newer versions. Details are explained in
`Documentation:
Installation `__.
.. code:: Bash
pip install --upgrade covsirphy
Demo
----
Quickest tour of CovsirPhy is here. The following codes analyze the
records in Japan.
.. code:: Python
import covsirphy as cs
# Data preparation,time-series segmentation, parameter estimation with SIR-F model
snr = cs.ODEScenario.auto_build(geo="Japan", model=cs.SIRFModel)
# Check actual records
snr.simulate(name=None);
# Show the result of time-series segmentation
snr.to_dynamics(name="Baseline").detect();
# Perform simulation with estimated ODE parameter values
snr.simulate(name="Baseline");
# Predict ODE parameter values (30 days from the last date of actual records)
snr.build_with_template(name="Predicted", template="Baseline");
snr.predict(days=30, name="Predicted");
# Perform simulation with estimated and predicted ODE parameter values
snr.simulate(name="Predicted");
# Add a future phase to the baseline (ODE parameters will not be changed)
snr.append();
# Show created phases and ODE parameter values
snr.summary()
# Compare reproduction number of scenarios (predicted/baseline)
snr.compare_param("Rt");
# Compare simulated number of cases
snr.compare_cases("Confirmed");
# Describe representative values
snr.describe()
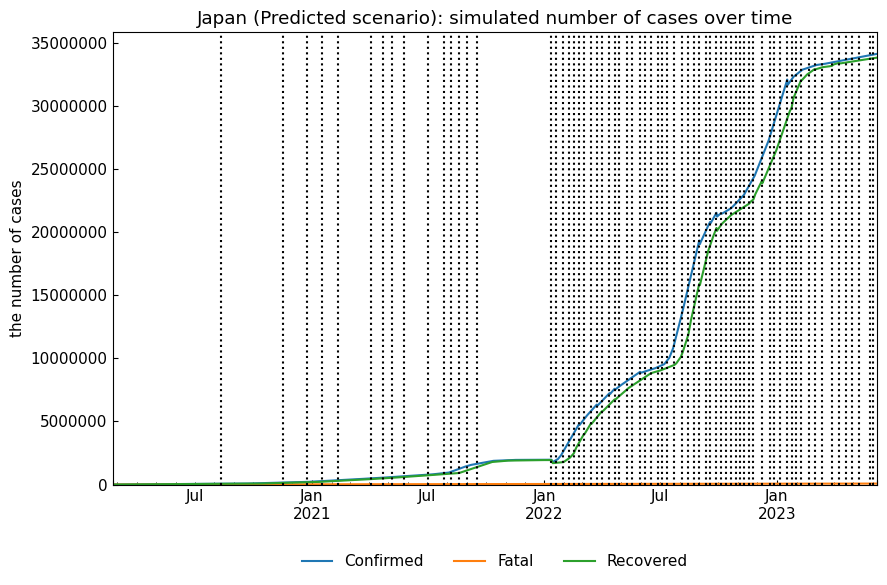
Output of ``snr.simulate(name="Predicted");``
.. raw:: html
Inspiration
-----------
- Monitor the spread of COVID-19/Monkeypox with SIR-derived ODE models
- Predict the number of cases in each country/province
- Find the relationship of reproductive number and measures taken by
each country
If you have ideas or need new functionalities, please join this project.
Any suggestions with `Github
Issues `__
and `Twitter: @lisphilar `__ are always
welcomed. Questions are also great. Please refer to `Guideline of
contribution `__.
Installation
------------
The latest stable version of CovsirPhy is available at `PyPI (The Python
Package Index): covsirphy `__ and
supports Python 3.8 or newer versions. Details are explained in
`Documentation:
Installation `__.
.. code:: Bash
pip install --upgrade covsirphy
Demo
----
Quickest tour of CovsirPhy is here. The following codes analyze the
records in Japan.
.. code:: Python
import covsirphy as cs
# Data preparation,time-series segmentation, parameter estimation with SIR-F model
snr = cs.ODEScenario.auto_build(geo="Japan", model=cs.SIRFModel)
# Check actual records
snr.simulate(name=None);
# Show the result of time-series segmentation
snr.to_dynamics(name="Baseline").detect();
# Perform simulation with estimated ODE parameter values
snr.simulate(name="Baseline");
# Predict ODE parameter values (30 days from the last date of actual records)
snr.build_with_template(name="Predicted", template="Baseline");
snr.predict(days=30, name="Predicted");
# Perform simulation with estimated and predicted ODE parameter values
snr.simulate(name="Predicted");
# Add a future phase to the baseline (ODE parameters will not be changed)
snr.append();
# Show created phases and ODE parameter values
snr.summary()
# Compare reproduction number of scenarios (predicted/baseline)
snr.compare_param("Rt");
# Compare simulated number of cases
snr.compare_cases("Confirmed");
# Describe representative values
snr.describe()
Output of ``snr.simulate(name="Predicted");``
.. raw:: html
 Tutorial
--------
Tutorials of functionalities are included in the `CovsirPhy
documentation `__.
- `Data
preparation `__
- `Data
Engineering `__
- `SIR-derived ODE
models `__
- `Phase-dependent SIR
models `__
- `Scenario
analysis `__
- `ODE parameter
prediction `__
Release notes
-------------
Release notes are
`here `__. Titles &
links of issues are listed with acknowledgement.
We can see the release plan for the next stable version in `milestone
page of the GitHub
repository `__. If
you find a highly urgent matter, please let us know via `issue
page `__.
Developers
----------
CovsirPhy library is developed by a community of volunteers. Please see
the full list
`here `__.
This project started in Kaggle platform. Hirokazu Takaya
(`@lisphilar `__) published `Kaggle
Notebook: COVID-19 data with SIR
model `__
on 12Feb2020 and developed it, discussing with Kaggle community. On
07May2020, "covid19-sir" repository was created. On 10May2020,
``covsirphy`` version 1.0.0 was published in GitHub. The first release
in PyPI (version 2.3.0) was on 28Jun2020. Many APIs were reviewed via
2.x series and version 3.0.0 was released on 12May2023.
Support
-------
Please support this project as a developer (or a backer). |Become a
backer|
License: Apache License 2.0
---------------------------
Please refer to
`LICENSE `__
file.
Citation
--------
Please cite this library as follows with version number
(``import covsirphy as cs; cs.__version__``).
**Hirokazu Takaya and CovsirPhy Development Team (2020-2024), CovsirPhy
version [version number]: Python library for COVID-19 analysis with
phase-dependent SIR-derived ODE
models,**\ https://github.com/lisphilar/covid19-sir
This is the output of ``covsirphy.__citation__``.
.. code:: Python
import covsirphy as cs
cs.__citation__
**We have no original papers the author and contributors wrote, but note
that some scientific approaches, including SIR-F model, S-R change point
analysis, phase-dependent approach to SIR-derived models, were developed
in this project.**
.. |PyPI version| image:: https://badge.fury.io/py/covsirphy.svg
:target: https://badge.fury.io/py/covsirphy
.. |Downloads| image:: https://static.pepy.tech/badge/covsirphy
:target: https://pepy.tech/project/covsirphy
.. |PyPI - Python Version| image:: https://img.shields.io/pypi/pyversions/covsirphy
:target: https://badge.fury.io/py/covsirphy
.. |GitHub license| image:: https://img.shields.io/github/license/lisphilar/covid19-sir
:target: https://github.com/lisphilar/covid19-sir/blob/master/LICENSE
.. |Quality Check| image:: https://github.com/lisphilar/covid19-sir/actions/workflows/test.yml/badge.svg
:target: https://github.com/lisphilar/covid19-sir/actions/workflows/test.yml
.. |Test Coverage| image:: https://codecov.io/gh/lisphilar/covid19-sir/branch/master/graph/badge.svg?token=9Z8Z1UHY3I
:target: https://codecov.io/gh/lisphilar/covid19-sir
.. |Become a backer| image:: https://opencollective.com/covsirphy/tiers/backer.svg?avatarHeight=36&width=600
:target: https://opencollective.com/covsirphy
Tutorial
--------
Tutorials of functionalities are included in the `CovsirPhy
documentation `__.
- `Data
preparation `__
- `Data
Engineering `__
- `SIR-derived ODE
models `__
- `Phase-dependent SIR
models `__
- `Scenario
analysis `__
- `ODE parameter
prediction `__
Release notes
-------------
Release notes are
`here `__. Titles &
links of issues are listed with acknowledgement.
We can see the release plan for the next stable version in `milestone
page of the GitHub
repository `__. If
you find a highly urgent matter, please let us know via `issue
page `__.
Developers
----------
CovsirPhy library is developed by a community of volunteers. Please see
the full list
`here `__.
This project started in Kaggle platform. Hirokazu Takaya
(`@lisphilar `__) published `Kaggle
Notebook: COVID-19 data with SIR
model `__
on 12Feb2020 and developed it, discussing with Kaggle community. On
07May2020, "covid19-sir" repository was created. On 10May2020,
``covsirphy`` version 1.0.0 was published in GitHub. The first release
in PyPI (version 2.3.0) was on 28Jun2020. Many APIs were reviewed via
2.x series and version 3.0.0 was released on 12May2023.
Support
-------
Please support this project as a developer (or a backer). |Become a
backer|
License: Apache License 2.0
---------------------------
Please refer to
`LICENSE `__
file.
Citation
--------
Please cite this library as follows with version number
(``import covsirphy as cs; cs.__version__``).
**Hirokazu Takaya and CovsirPhy Development Team (2020-2024), CovsirPhy
version [version number]: Python library for COVID-19 analysis with
phase-dependent SIR-derived ODE
models,**\ https://github.com/lisphilar/covid19-sir
This is the output of ``covsirphy.__citation__``.
.. code:: Python
import covsirphy as cs
cs.__citation__
**We have no original papers the author and contributors wrote, but note
that some scientific approaches, including SIR-F model, S-R change point
analysis, phase-dependent approach to SIR-derived models, were developed
in this project.**
.. |PyPI version| image:: https://badge.fury.io/py/covsirphy.svg
:target: https://badge.fury.io/py/covsirphy
.. |Downloads| image:: https://static.pepy.tech/badge/covsirphy
:target: https://pepy.tech/project/covsirphy
.. |PyPI - Python Version| image:: https://img.shields.io/pypi/pyversions/covsirphy
:target: https://badge.fury.io/py/covsirphy
.. |GitHub license| image:: https://img.shields.io/github/license/lisphilar/covid19-sir
:target: https://github.com/lisphilar/covid19-sir/blob/master/LICENSE
.. |Quality Check| image:: https://github.com/lisphilar/covid19-sir/actions/workflows/test.yml/badge.svg
:target: https://github.com/lisphilar/covid19-sir/actions/workflows/test.yml
.. |Test Coverage| image:: https://codecov.io/gh/lisphilar/covid19-sir/branch/master/graph/badge.svg?token=9Z8Z1UHY3I
:target: https://codecov.io/gh/lisphilar/covid19-sir
.. |Become a backer| image:: https://opencollective.com/covsirphy/tiers/backer.svg?avatarHeight=36&width=600
:target: https://opencollective.com/covsirphy
 Inspiration
-----------
- Monitor the spread of COVID-19/Monkeypox with SIR-derived ODE models
- Predict the number of cases in each country/province
- Find the relationship of reproductive number and measures taken by
each country
If you have ideas or need new functionalities, please join this project.
Any suggestions with `Github
Issues
Inspiration
-----------
- Monitor the spread of COVID-19/Monkeypox with SIR-derived ODE models
- Predict the number of cases in each country/province
- Find the relationship of reproductive number and measures taken by
each country
If you have ideas or need new functionalities, please join this project.
Any suggestions with `Github
Issues  Tutorial
--------
Tutorials of functionalities are included in the `CovsirPhy
documentation
Tutorial
--------
Tutorials of functionalities are included in the `CovsirPhy
documentation  |PyPI version| |Downloads| |PyPI - Python Version| |GitHub license|
|Quality Check| |Test Coverage|
CovsirPhy introduction
======================
`Documentation
|PyPI version| |Downloads| |PyPI - Python Version| |GitHub license|
|Quality Check| |Test Coverage|
CovsirPhy introduction
======================
`Documentation